Xu Fei, Jiangnan University, Biomacromolecules: design strategy for adjusting the structure and mechanical properties of synthetic collagen hydrogels
[Abstract]
as an important part of biomaterials, collagen provides three-dimensional scaffolds and biological clues for cell adhesion and proliferation in tissue engineering. The recombinant collagen like protein, which was originally found in Streptococcus pyogenes and produced in heterologous hosts, has been chemically and genetically engineered for biomaterial applications. However, existing collagen like proteins do not form gels, limiting their usefulness as biomaterials.
Recently, Professor Xu Fei s team of Jiangnan University proposed a series of reasonably designed collagen like domains, which are composed of trimerization domains, three helix domains with different lengths and a pair of collagen like domains connected to the membrane as adhesive terminals The N-terminal and C-terminal heterotrimers are composed of coiled coil sequences. These designed proteins fold into three helices and form self supporting gels. With the extension of the three helix domain, the gel becomes less rigid, the pore size increases, and the structural anisotropy decreases. In addition, cell culture experiments confirmed that the designed protein was non cytotoxic. This study provides a design strategy for collagen based biomaterials. Sequence changes reveal the relationship between protein primary structure and material properties, and the change of crosslinking density and binding energy defines gelation of protein network. The related papers were published in biomacromolecules with the title of design strategies to tune the structural and mechanical properties of synthetic collagen hydrogels.
[main picture guide]
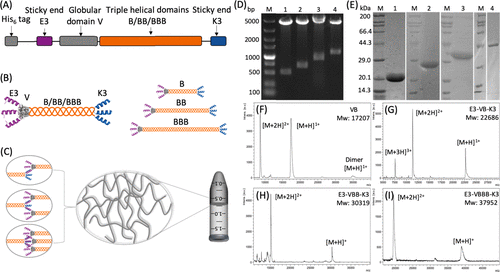
chart 1. Protein design strategy and validation. Three designs were tested: e3-vb-k3, e3-vbb-k3 and e3-vbbb-k3. The schematic diagram of (a) sequence and (b) secondary structure of protein( C) The proposed intermolecular association and self-assembly mechanism. Gel electrophoresis (D) was used to digest the length of the nucleotide fragment and (E) the molecular weight of the recombinant plasmid with NcoI and BamHI enzymes. Matrix assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry was used to characterize the molecular weight of proteins.
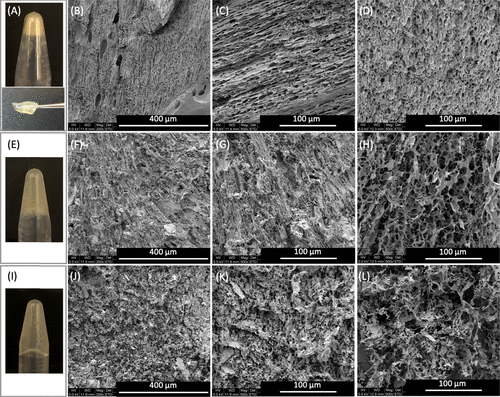
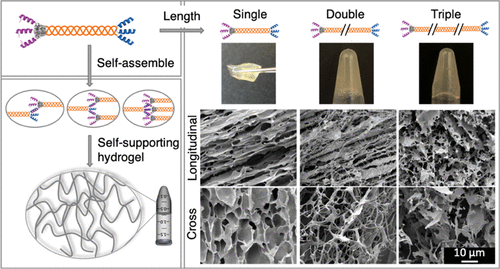
chart 2. gel structure. The gelation of A-D) E3-VB-K3 was tested by inverted vial and tweezers operation. After freeze-drying, the microstructure of the gel was cut into (B, C) longitudinal and (D) cross sections and observed by scanning electron microscope (SEM). E-H) e3-vbb-k3, with (F, g) longitudinal and (H) cross sections, and (I-L) e3-vbbb-k3, with (J, K) longitudinal and (L) transverse cross sections. Because of the low hardness of E3-VBB-K3 and E3-VBBB-K3 gels, they can not be treated with tweezers.
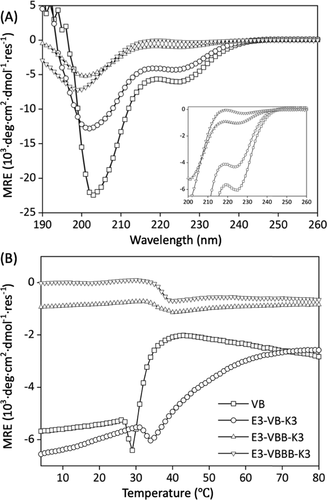
chart 3. Protein structure and stability( A) CD wavelength scanning and thermal denaturation curves of (b) VB, e3-vb-k3, e3-vbb-k3 and e3-vbbb-k3 at 1 mg / ml in 10 mM phosphate buffer (pH 7.4) were performed.
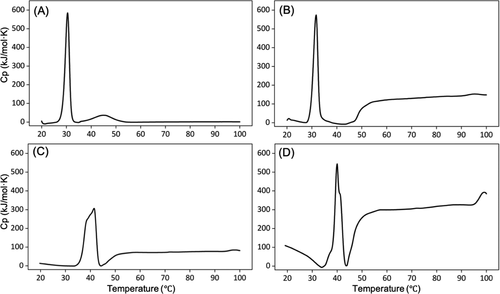
chart four The designed collagen (a) VB, (b) e3-vb-k3, (c) e3-vbb-k3 and (d) e3-vbbb-k3 with the concentration of 10 mg / ml in 10 mm were determined by differential scanning calorimetry phosphate buffer (pH 7.4).
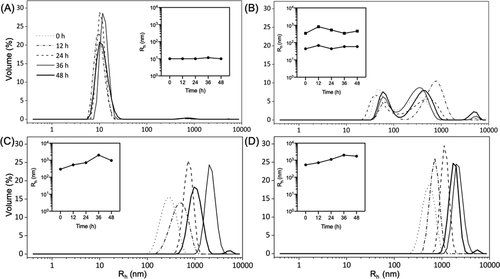
chart 5. Particle size distribution. Dynamic light scattering (DLS) measurements of the hydrodynamic radius (RH) distribution of collagen (a) VB, (b) e3-vb-k3, (c) e3-vbb-k3 and (d) e3-vbbb - were performed for more than 48 hours. The sample was kept at 4 ° C in 10 mM phosphate buffer at a concentration of about 1 mg / ml (pH 7.4). The illustration of each subgraph shows the change of the main RH peak (> 5% volume intensity) with time. In all subgraphs, the RH distribution curves at different times are shown in the right figure.
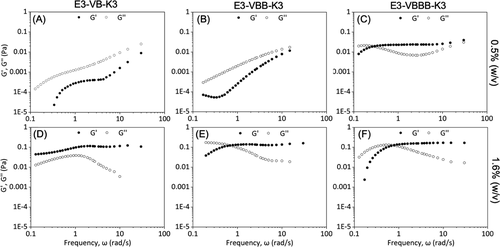
chart 6. Micro rheology( A. D) e3-vb-k3, (B, e) e3-vbb-k3 and (C, f) e3-vbbb-k3 protein solutions at 0.5 and 1.6% (w / V) in 10 mM phosphate buffer (pH 7.4). The concentration of e3-vbbb-k3 (f) was 1.16% (w / V).
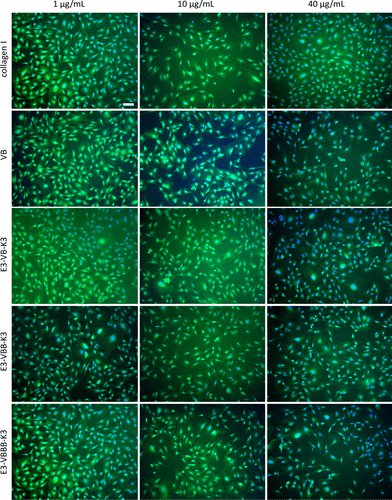
chart 7. The cells were spread on the plate coated with designed collagen. NIH / 3T3 mouse fibroblasts were seeded and spread on tissue culture plates coated with natural type I collagen (positive control), VB, e3-vb-k3, e3-vbb-k3 and e3-vbbb-k3 at concentrations of 1, 10 and 40 μ G / ml for 24 hours. The protein name and concentration are labeled on the left and top of the graph, respectively. The cells were stained with CFDA Se and Hoechst and observed with fluorescence microscope. The scale represents 100 of all images μ m。
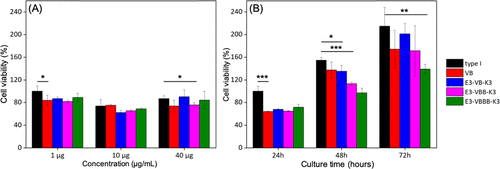
chart eight Relative cell viability on substrates composed of designed proteins of different (a) concentrations and (b) incubation times. The sample name is on the right. Natural type I collagen (black) was tested as positive control.
[Conclusion] the team developed recombinant collagen that was self organized into gel by protein design. Use collagen to connect to Construction of scl2 fragment α- The large-scale and low-cost production of protein materials is possible. Compared with natural collagen, these designed proteins have several advantages, such as consistency between batches, easy genetic manipulation, and modular design for carrying sequences endowed with biological functions, such as integrin binding. Cell culture experiments confirmed that the gel was cytotoxic and could be used for three-dimensional cell culture.
The structural and mechanical properties of cell environment affect cell viability and fate. By modifying the protein primary sequence, the team can adjust the material properties in the chimeric design, such as pore size, anisotropy of microstructure, gel strength and moisture content. E3-VB-K3 forms a hard gel with anisotropic microstructure, composed of uniform pores. E3-VBBB-K3 formed homogeneous hydrogel with more porous microstructure and high (> 98%) water content. In addition, the observed anisotropic microstructures are usually related to the sequential assembly process of polymers induced by intermolecular interactions and external forces such as magnetic field and mechanical stretching. The team s collagen with an elongated triple helix domain seems to increase the probability of collision, accelerate the assembly process and destroy the uniaxial orientation of molecules. stay In DLS, e3-vb-k3 formed stable small particles and medium size (~ 60 and ~ 500 nm) with time, while the particle size of e3-vbbb-k3 increased from ~ 300 to ~ 3000 nm.
This information is from the Internet for academic exchange only. If there is any infringement, please contact us to delete it immediately
18915694570
Previous: AFM by Mei Yingwu of Z


